Background
The ACRF Cancer Genomics Facility is a fee for service core facility located within the Centre for Cancer Biology (SA Pathology). An extensive list of equipment available to the South Australian research community can be found on our website along with contact information for members of the Facility. Please note that although our primary focus is cancer research we will consider other projects on a case by case basis.
Objective
The ACRF Cancer Genomics Facility provides a number of microarray and sequencing services for a variety of applications. This guide is intended to answer common questions regarding sample submission and starting material requirements for the different technologies. If you have low quantity or low concentration samples, please contact a member of the Facility and we will do our best to work with what you have.
New Project Requests
Please make sure you have discussed your project with us before sending samples. After contacting us, a member of the Facility will send you a quote that clearly outlines the cost of the project including estimated sequencing read numbers, data output and/or expected sequencing coverage. When submitting samples please fill out the attached excel spreadsheet with relevant sample information and email it to us.
Download the sample submission form
Sample Purification
A number of methods/kits are available for the isolation of total RNA and/or high molecular weight DNA from cells and tissue. Kits from Roche, Qiagen and Life Technologies perform well. If performing organic extractions take extra care to avoid carry over of organics that can inhibit downstream enzymatic reactions. Including a DNase digest in the isolation of total RNA samples is recommended, particularly for RNA sequencing applications. Keep in mind that traditional total RNA isolation methods (Trizol, RNeasy columns, etc) tend to lose small RNAs. For the isolation of total RNA containing small RNAs or for the enrichment of small RNAs, the mirVana isolation kit (Part No. AM1560 or AM1561) from Thermo Fisher (formerly Ambion) is highly recommended. Store and/or dilute both DNA and RNA in molecular biology grade water or low TE with EDTA 0.1mM or less.
Sample Quality Control
The ACRF Cancer Genomics Facility utilizes a number of different technologies for sample quality control including the Bioanalyzer, Qubit 2.0 Fluorometer, Shimadzu MultiNA, Trinean DropSense, the Nanodrop as well as qPCR. At a minimum, all samples submitted to the Facility will be run on the appropriate Bioanalyzer chip except for ChIP DNA and genomic DNA. Genomic DNA should be run on a 1% agarose gel prior to submission to confirm integrity and/or lack of RNA contamination. For ChIP samples, relative enrichment should be confirmed with a control qPCR assay comparing the input sample and IP sample. The recommendations in this section apply to samples that will ONLY be run on the Bioanalyzer. If submitting samples for microarrays or sequencing, the amount of sample required for the Bioanalyzer has already been accounted for in subsequent sections.
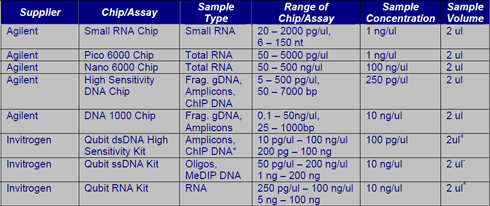
*ChIP DNA samples typically have concentrations less than 10 ng/ul and are often undetected by the Nanodrop and Bioanalyzer. We recommend using the Qubit dsDNA High Sensitivity Kit to determine the concentration of ChIP samples. This picogreen assay is highly selective for double-stranded DNA (dsDNA) over ssDNA, RNA or other contaminants.
+ At least 200 pg should enter the assay. Additional volume may be required if the sample concentration is < 100 pg/ul
- At least 1ng should enter the assay. Additional volume may be required is sample concentration is < 1ng/ul
^ At least 5ng should enter the assay. Additional volume may be required is sample concentration is < 5ng/ul
The ACRF Cancer Genomics Facility will notify researchers of any total RNA samples having RINs less than 7.0 to allow for replacement of the samples or to consider a different experimental approach. Also note that the RIN calculation assumes the presence of 18 and 28s ribosomal bands. If your RNA has been isolated from organisms other than human, mouse and rat, etc. please let us know accordingly.
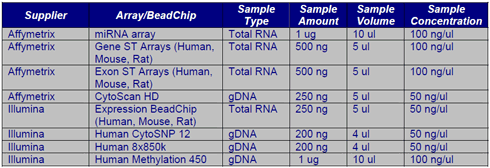
Microarrays
The ACRF Cancer Genomics Facility has the equipment to run both Affymetrix microarray and Illumina BeadArray experiments. Sample requirements for the common array designs and applications are listed below.
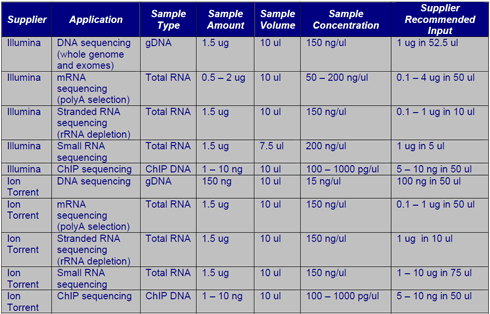
Next-Generation Sequencing
The ACRF Cancer Genomics Facility offers library preparation and sequencing services for Illumina (HiSeq 2500, NextSeq and MiSeq) and Ion Torrent (PGM and Proton). Refer to your application of interest for sample input and volume recommendations. If you’re struggling to come up with the recommended amounts contact a member of the Genomics Facility for other options. Also note that Exome sequencing protocols begin with standard gDNA library preps, hence 1.5 µgs of gDNA is required for Exome sequencing.
Fluidigm BioMark HD and Access Array
The ACRF Cancer Genomics Facility offers high-throughput qPCR Genotyping and Gene Expression assays on the Fluidigm BioMark HD. The Fluidigm workflow is compatible with standard TaqMan chemistry, Fluidigm Assays or custom primers with EvaGreen (SybrGreen equivalent). We run this as a service, but are also willing to train users how to use the technology. Fluidigm currently has 4 chip options that can accommodate a number of sample and assay combinations. See table below for more information. As with most technologies more reactions leads to a lower price per reaction.
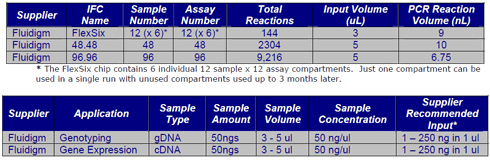
Samples and primers should be provided into 96-well plates or 0.2mL 8-tube strips arranged in columns. If STA is required a pool of primers should also be supplied in a 1.5mL tube.
The Fluidigm Access Array enables high-throughput amplicon generation for next-generation sequencing. Up to 384 barcodes are available for high level sample multiplexing during a sequencing run. The Access Array contains 48 sample wells by 48 primer wells. Up to 10 primer pairs can be multiplexed in each primer well. You can design primers for regions of interest or have Fluidigm design the assays for you.

Sequenom MassARRAY mass spectrometry (Agena Bioscience)
The ACRF Cancer Genomics Facility offers high-throughput, scalable, cost-effective, accurate and sensitive genotyping and somatic mutation detection using Sequenom MassArray MALDO-TOF mass spectrometry. We can run this as a service and provide you with results in a spreadsheet or we can train users to use the technology. Users can design custom assays using the online design tool, purchase pre-designed panels, have the Agena Custom Services Laboratory design the assays, or you can contact a member of our facility of assay design help. Generally, up to 40 SNPs or 10 somatic mutations can be multiplexed and examined in a single reaction. At least 10 ng of DNA per reaction is recommended by the manufacturer (gDNA, cDNA or amplicon, depending on the application). Samples should be supplied in a 96- or 384-well plate. Reactions are performed in a 384-well plate.




